Welcome to ProteinShake!
Protein structure datasets and tasks in any learning framework… in one line.
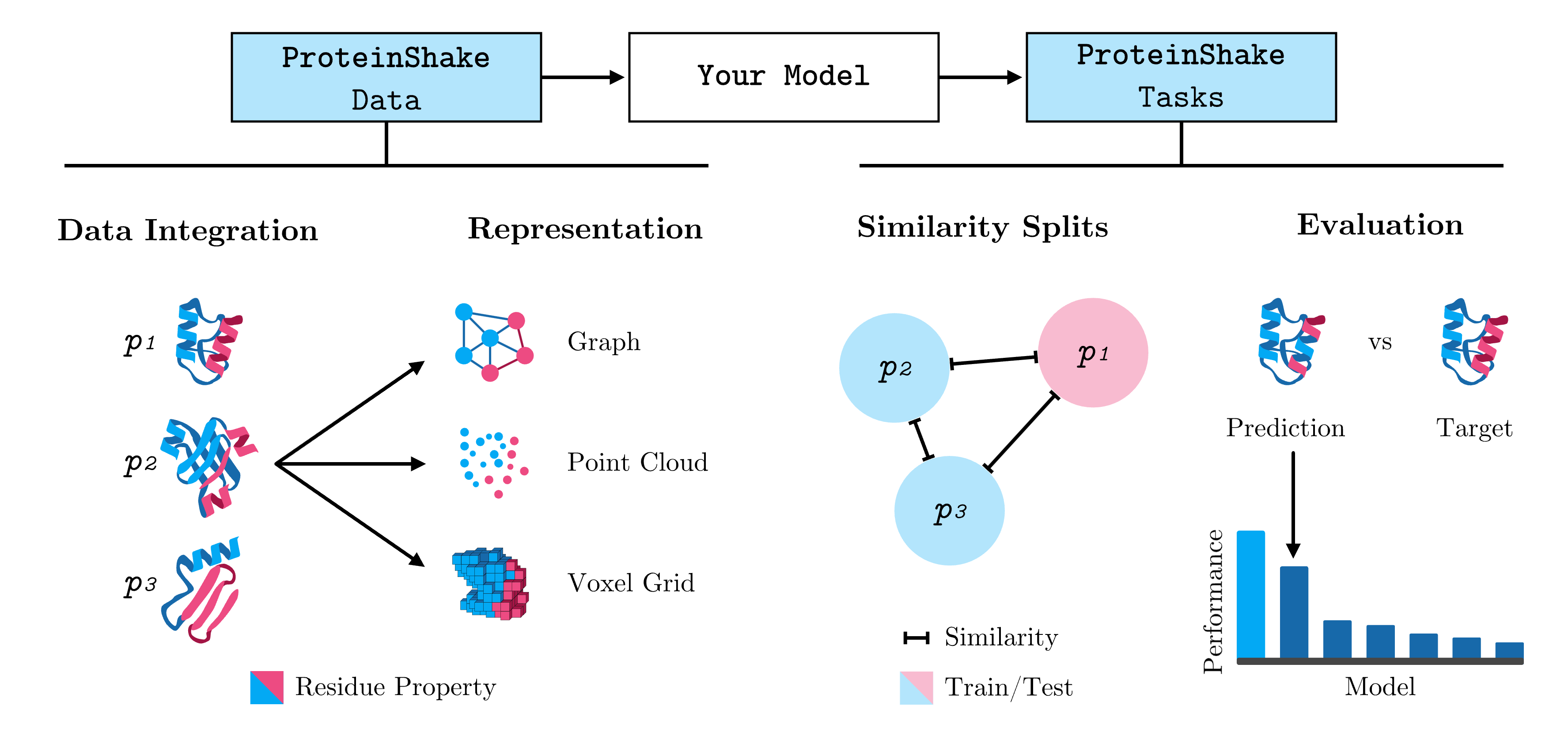
We provide a collection of pre-processed and cleaned protein 3D structure datasets from RCSB and AlphaFoldDB, including annotations. Structures are easily converted to graphs, voxels, or point clouds and loaded natively into PyTorch, TensorFlow, NumPy, JAX, PyTorch Geometric, DGL and NetworkX. The task API enables standardized benchmarking on a variety of tasks on protein and residue level.
Note
Check out the Installation Guide, the Quickstart, or our Website to get started.
Note
We welcome contributions and bug reports through issues and pull requests on GitHub. See also our Contribution Guide.
Who is ProteinShake for?
ProteinShake is intended for computational biologists and machine learning researchers who need accessible datasets and well-defined evaluation benchmarks for their deep learning models.
We put emphasis on extendability, aiming to eliminate boilerplate code for data preparation and model evaluation across machine learning disciplines. ProteinShake therefore also serves as a general framework for processing protein structure data, and we hope it will serve the community as a platform to share their datasets and evaluation tasks. New datasets and tasks can be created with just a few lines of code (see the Tutorial) and we will integrate your contributions through pull requests on GitHub (see the Contribution Guide).
Notes
Package Reference